Filter pipelines¶
This example shows how to use the pymia.filtering package to set up image filter pipeline and apply it to an image. The pipeline consists of a gradient anisotropic diffusion filter followed by a histogram matching. This pipeline will be applied to a T1-weighted MR image and a T2-weighted MR image will be used as a reference for the histogram matching.
Tip
This example is available as Jupyter notebook at ./examples/filtering/basic.ipynb and Python script at ./examples/filtering/basic.py.
Note
To be able to run this example:
Get the example data by executing ./examples/example-data/pull_example_data.py.
Install matplotlib (
pip install matplotlib).
Import the required modules.
[1]:
import glob
import os
import matplotlib.pyplot as plt
import pymia.filtering.filter as flt
import pymia.filtering.preprocessing as prep
import SimpleITK as sitk
Define the path to the data.
[2]:
data_dir = '../example-data'
Let us create a list with the two filters, a gradient anisotropic diffusion filter followed by a histogram matching.
[3]:
filters = [
prep.GradientAnisotropicDiffusion(time_step=0.0625),
prep.HistogramMatcher()
]
histogram_matching_filter_idx = 1 # we need the index later to update the HistogramMatcher's parameters
Now, we can initialize the filter pipeline.
[4]:
pipeline = flt.FilterPipeline(filters)
We can now loop over the subjects of the example data. We will both load the T1-weighted and T2-weighted MR images and execute the pipeline on the T1-weighted MR image. Note that for each subject, we update the parameters for the histogram matching filter to be the corresponding T2-weighted image.
[5]:
# get subjects to evaluate
subject_dirs = [subject for subject in glob.glob(os.path.join(data_dir, '*')) if os.path.isdir(subject) and os.path.basename(subject).startswith('Subject')]
for subject_dir in subject_dirs:
subject_id = os.path.basename(subject_dir)
print(f'Filtering {subject_id}...')
# load the T1- and T2-weighted MR images
t1_image = sitk.ReadImage(os.path.join(subject_dir, f'{subject_id}_T1.mha'))
t2_image = sitk.ReadImage(os.path.join(subject_dir, f'{subject_id}_T2.mha'))
# set the T2-weighted MR image as reference for the histogram matching
pipeline.set_param(prep.HistogramMatcherParams(t2_image), histogram_matching_filter_idx)
# execute filtering pipeline on the T1-weighted image
filtered_t1_image = pipeline.execute(t1_image)
# plot filtering result
slice_no_for_plot = t1_image.GetSize()[2] // 2
fig, axs = plt.subplots(1, 2)
axs[0].imshow(sitk.GetArrayFromImage(t1_image[:, :, slice_no_for_plot]), cmap='gray')
axs[0].set_title('Original image')
axs[1].imshow(sitk.GetArrayFromImage(filtered_t1_image[:, :, slice_no_for_plot]), cmap='gray')
axs[1].set_title('Filtered image')
fig.suptitle(f'{subject_id}', fontsize=16)
plt.show()
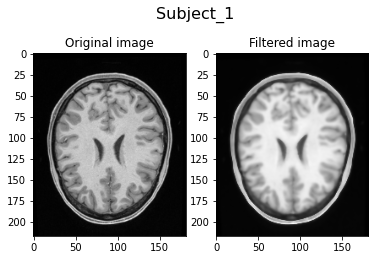
Filtering Subject_1...
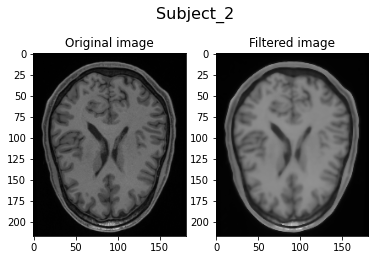
Filtering Subject_2...
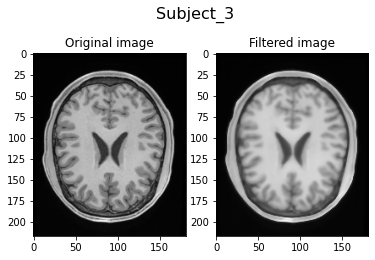
Filtering Subject_3...
Filtering Subject_4...
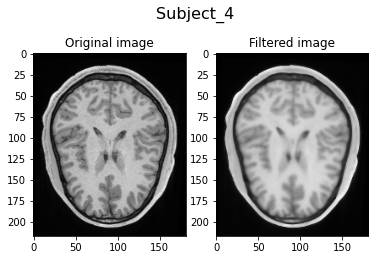
Visually, we can clearly see the smoothing of the filtered image due to the anisotrophic filtering. Also, the image intensities are brighter due to the histogram matching.