Creation of a dataset¶
This example shows how to use the pymia.data package to create a HDF5 (hierarchical data format version 5) dataset. All examples follow the use case of medical image segmentation of brain tissues, see Examples for an introduction into the data. Therefore, we create a dataset with the four subjects and their data: a T1-weighted MR image, a T2-weighted MR image, a label image (ground truth, GT), and a mask image, as well as demographic information age, grade point average
(GPA), and gender.
Tip
This example is available as Jupyter notebook at ./examples/data/creation.ipynb and Python script at ./examples/data/creation.py.
Note
To be able to run this example:
Get the example data by executing ./examples/example-data/pull_example_data.py.
Import the required modules.
[1]:
import enum
import glob
import os
import typing
import SimpleITK as sitk
import numpy as np
import pymia.data as data
import pymia.data.conversion as conv
import pymia.data.definition as defs
import pymia.data.creation as crt
import pymia.data.transformation as tfm
import pymia.data.creation.fileloader as file_load
Let us first define an enumeration with the data we will write to the dataset.
[2]:
class FileTypes(enum.Enum):
T1 = 1 # The T1-weighted MR image
T2 = 2 # The T2-weighted MR image
GT = 3 # The label (ground truth) image
MASK = 4 # The foreground mask
AGE = 5 # The age
GPA = 6 # The GPA
GENDER = 7 # The gender
Next, we define a subject. Each subject will have two structural MR images (T1w, T2w), one label image (ground truth), a mask, two numericals (age and GPA), and the gender (a character “m” or “w”).
[3]:
class Subject(data.SubjectFile):
def __init__(self, subject: str, files: dict):
super().__init__(subject,
images={FileTypes.T1.name: files[FileTypes.T1], FileTypes.T2.name: files[FileTypes.T2]},
labels={FileTypes.GT.name: files[FileTypes.GT]},
mask={FileTypes.MASK.name: files[FileTypes.MASK]},
numerical={FileTypes.AGE.name: files[FileTypes.AGE], FileTypes.GPA.name: files[FileTypes.GPA]},
gender={FileTypes.GENDER.name: files[FileTypes.GENDER]})
self.subject_path = files.get(subject, '')
We now collect the subjects, and initialize a Subject holding paths to each of the data.
[4]:
data_dir = '../example-data'
# get subjects
subject_dirs = [subject_dir for subject_dir in glob.glob(os.path.join(data_dir, '*')) if os.path.isdir(subject_dir) and os.path.basename(subject_dir).startswith('Subject')]
sorted(subject_dirs)
# the keys of the data to write to the dataset
keys = [FileTypes.T1, FileTypes.T2, FileTypes.GT, FileTypes.MASK, FileTypes.AGE, FileTypes.GPA, FileTypes.GENDER]
subjects = []
# for each subject on file system, initialize a Subject object
for subject_dir in subject_dirs:
id_ = os.path.basename(subject_dir)
file_dict = {id_: subject_dir} # init dict with id_ pointing to the path of the subject
for file_key in keys:
if file_key == FileTypes.T1:
file_name = f'{id_}_T1.mha'
elif file_key == FileTypes.T2:
file_name = f'{id_}_T2.mha'
elif file_key == FileTypes.GT:
file_name = f'{id_}_GT.mha'
elif file_key == FileTypes.MASK:
file_name = f'{id_}_MASK.nii.gz'
elif file_key == FileTypes.AGE or file_key == FileTypes.GPA or file_key == FileTypes.GENDER:
file_name = f'{id_}_demographic.txt'
else:
raise ValueError('Unknown key')
file_dict[file_key] = os.path.join(subject_dir, file_name)
subjects.append(Subject(id_, file_dict))
Then, we define a LoadData class. We load the structural MR images (T1w and T2w) as float and the other images as int. The age, GPA, and gender are loaded from the text file.
[5]:
class LoadData(file_load.Load):
def __call__(self, file_name: str, id_: str, category: str, subject_id: str) -> \
typing.Tuple[np.ndarray, typing.Union[conv.ImageProperties, None]]:
if id_ == FileTypes.AGE.name:
with open(file_name, 'r') as f:
value = np.asarray([int(f.readline().split(':')[1].strip())])
return value, None
if id_ == FileTypes.GPA.name:
with open(file_name, 'r') as f:
value = np.asarray([float(f.readlines()[1].split(':')[1].strip())])
return value, None
if id_ == FileTypes.GENDER.name:
with open(file_name, 'r') as f:
value = np.array(f.readlines()[2].split(':')[1].strip())
return value, None
if category == defs.KEY_IMAGES:
img = sitk.ReadImage(file_name, sitk.sitkFloat32)
else:
# this is the ground truth (defs.KEY_LABELS) and mask, which will be loaded as unsigned integer
img = sitk.ReadImage(file_name, sitk.sitkUInt8)
# return both the image intensities as np.ndarray and the properties of the image
return sitk.GetArrayFromImage(img), conv.ImageProperties(img)
Finally, we can use a writer to create the HDF5 dataset by passing the list of Subjects and the LoadData to a Traverser. For the structural MR images, we also apply an intensity normalization.
[6]:
hdf_file = '../example-data/example-dataset.h5'
# remove the "old" dataset if it exists
if os.path.exists(hdf_file):
os.remove(hdf_file)
with crt.get_writer(hdf_file) as writer:
# initialize the callbacks that will actually write the data to the dataset file
callbacks = crt.get_default_callbacks(writer)
# add a transform to normalize the structural MR images
transform = tfm.IntensityNormalization(loop_axis=3, entries=(defs.KEY_IMAGES, ))
# run through the subject files (loads them, applies transformations, and calls the callback for writing them)
traverser = crt.Traverser()
traverser.traverse(subjects, callback=callbacks, load=LoadData(), transform=transform)
start dataset creation
[1/4] Subject_1
[2/4] Subject_2
[3/4] Subject_3
[4/4] Subject_4
dataset creation finished
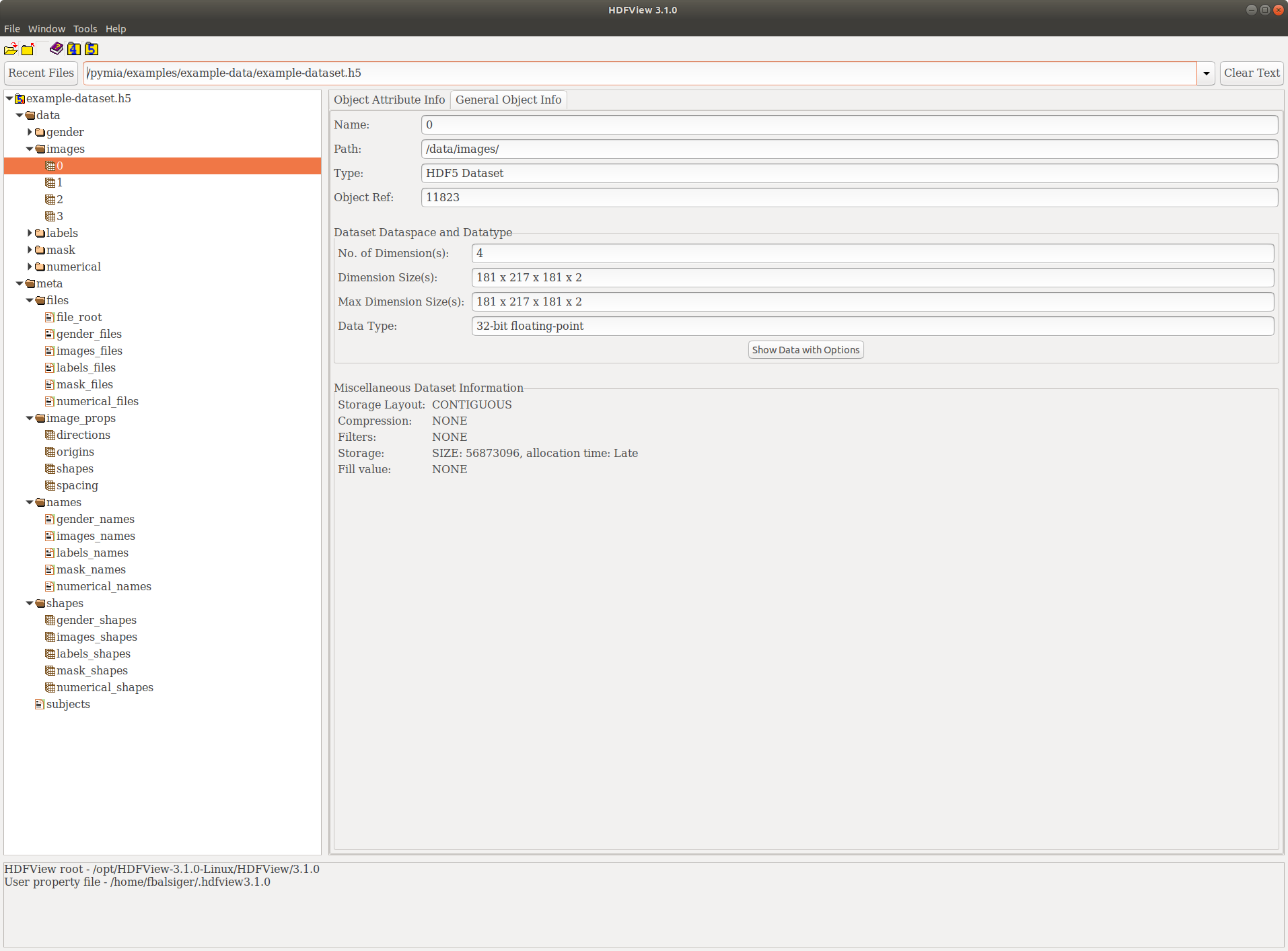
This should now have created a example-dataset.h5 in the directory ./examples/example-data. By using a HDF5 viewer like HDF Compass or HDFView, we can inspect the dataset. It should look similar to the figure below.